Yay! At this rate you guys will need to throw many parties in the lab.
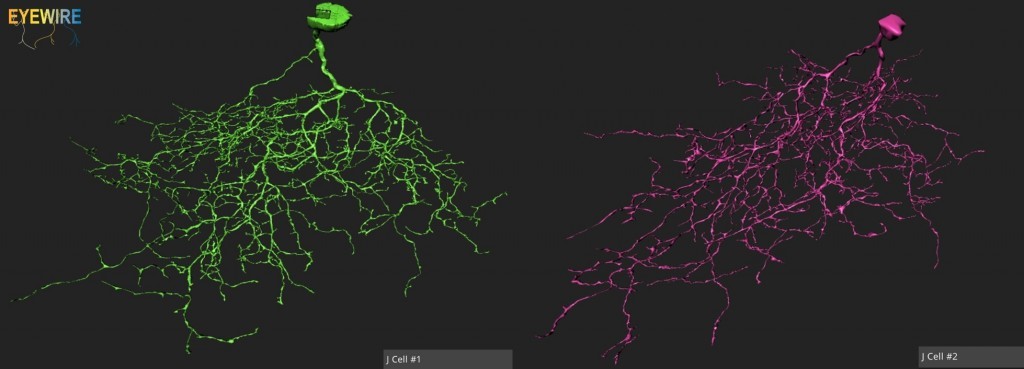
Before I answer your question, I’ve a question of my own: how come the transgenic mice were not used? It seems like it would have been useful to be able to determine the location of J-cells in order to make it easier to positively identify them, as well as map their distribution. Also, it would enable the comparison of the morphology of the symmetric J-cells and the asymmetric J-cells. I do not think it would be that hard (though I may be wrong) to combine serial block face-scanning electron microscopy with a method for identifying where the J-cells are. Just pass the slices from the ultramicrotome (or the blocks? Not sure what the configuration of the EM and the ultramicrotome are, so I don’t know which would easier to do in terms of space) under a photometer with a UV filter and a UV light source to measure the amount of YFP in the slice by the amount of light produced via fluorescence, and then construct a volumetric map of the resulting data, which would enable you to locate which cubes containing the cell bodies/neurites of the J-cells.