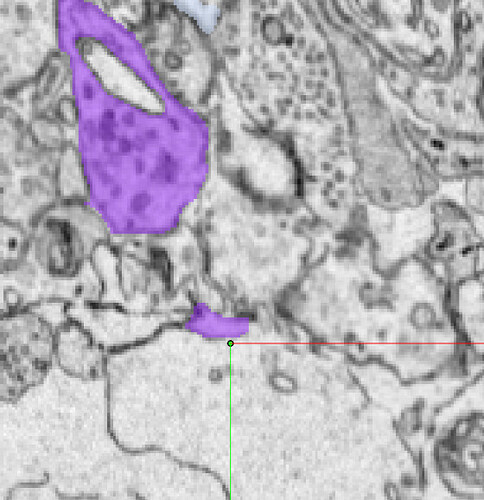
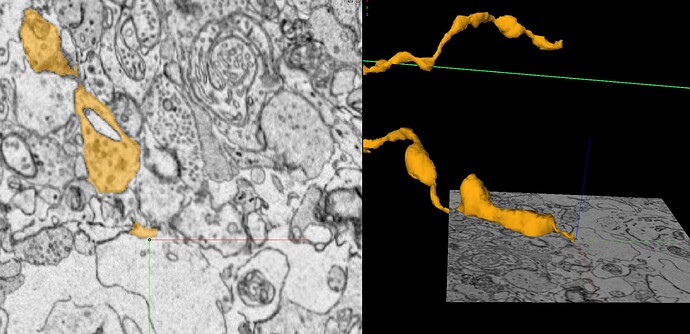
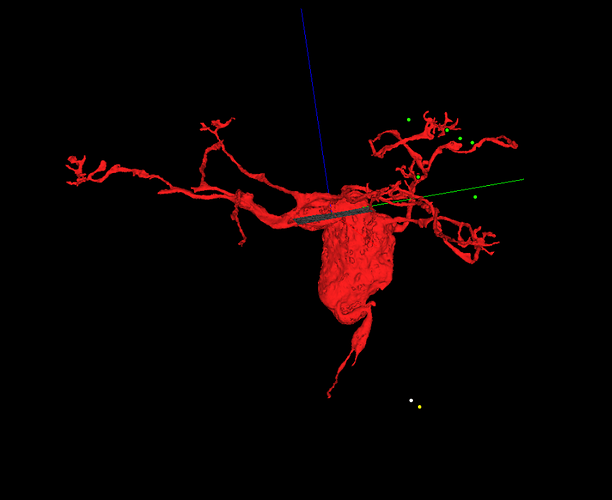
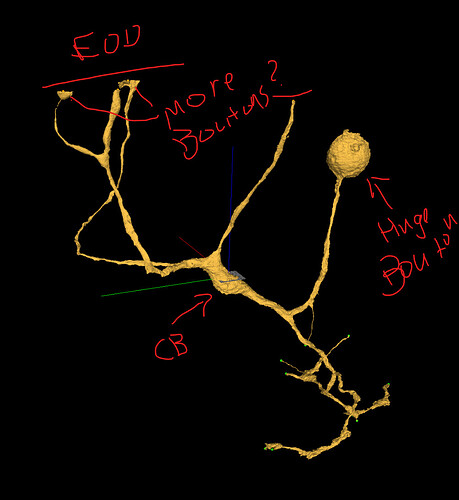
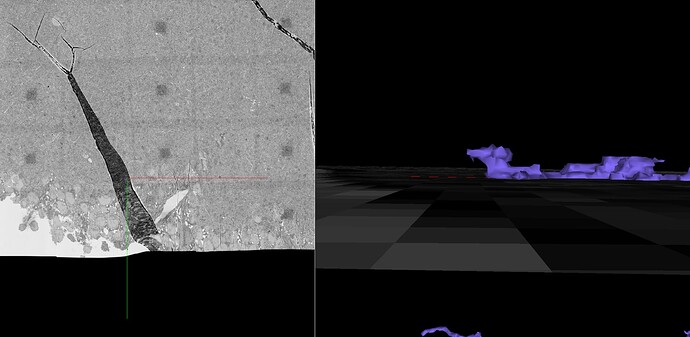
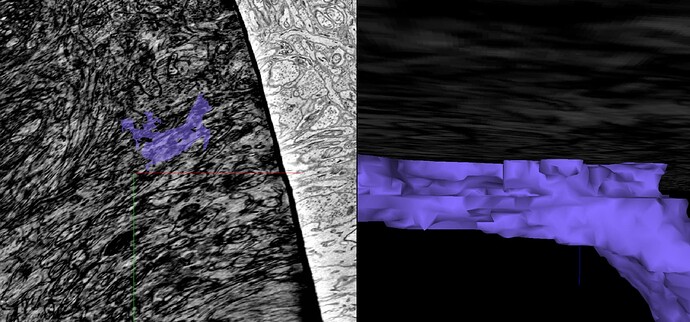
Unfortunately, the supervoxels you see are the smallest segments available in the segmentation. So we sort of just have to go with leaving the supervoxel with what branch makes the most sense and leave gaps like we had to in Eyewire.
The MSTY threshold slider was to control the association of the supervoxels to each other. So the higher affinity threshold meant more segments/supervoxels were connected together by the system and the lowest affinity threshold was the max supervoxels/segments we could break down into individual pieces.
For EW2, we decided to go with a slightly higher affinity threshold for the dataset overall because it’s easier to spot mergers than missing segments. However, ng allows us to be able to cut/view the supervoxels (smallest segmentation) at the same time in the 2D. So we don’t have to worry about sliding the threshold slider up and down like we had to in MSTY.


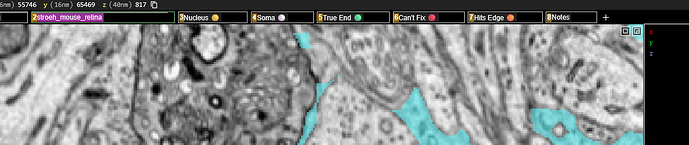
 ).
).